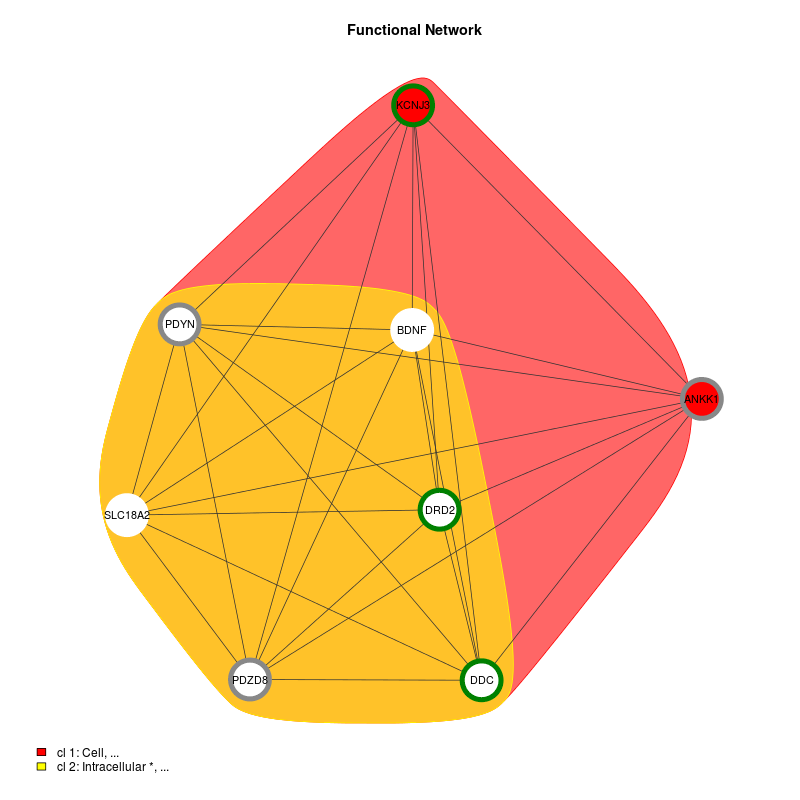
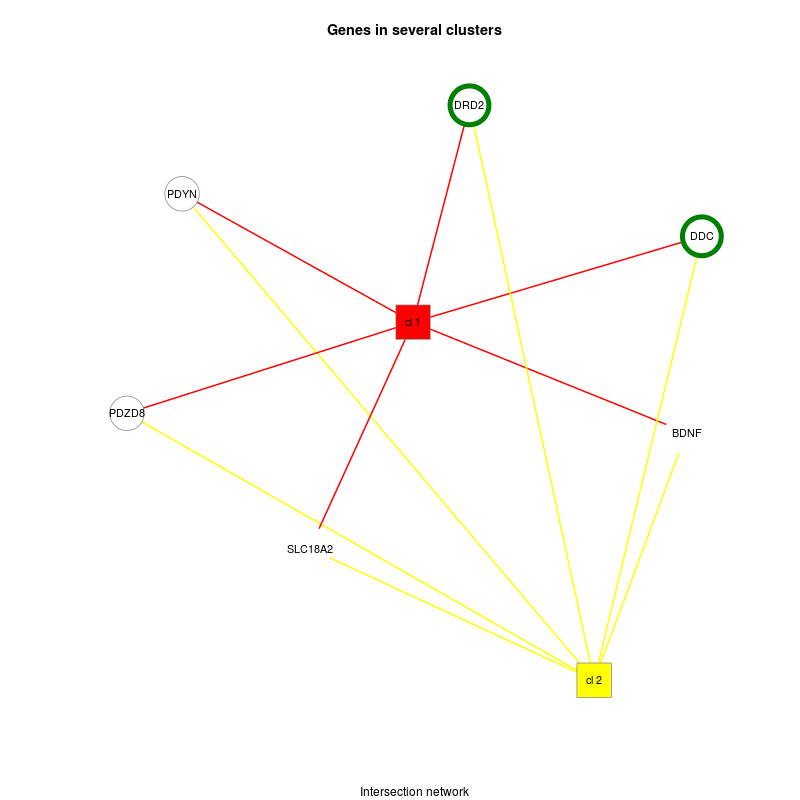
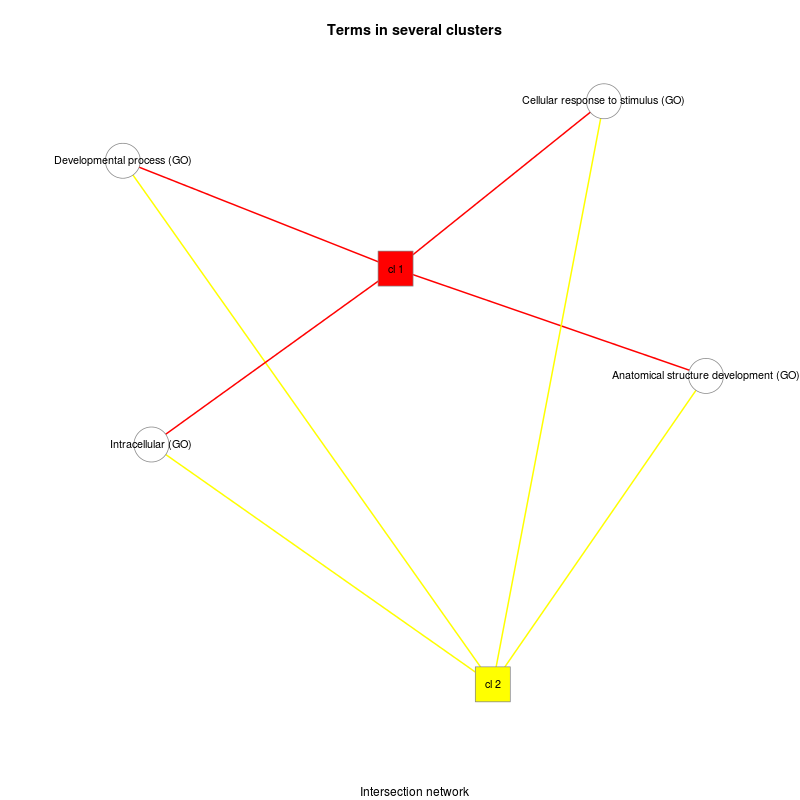
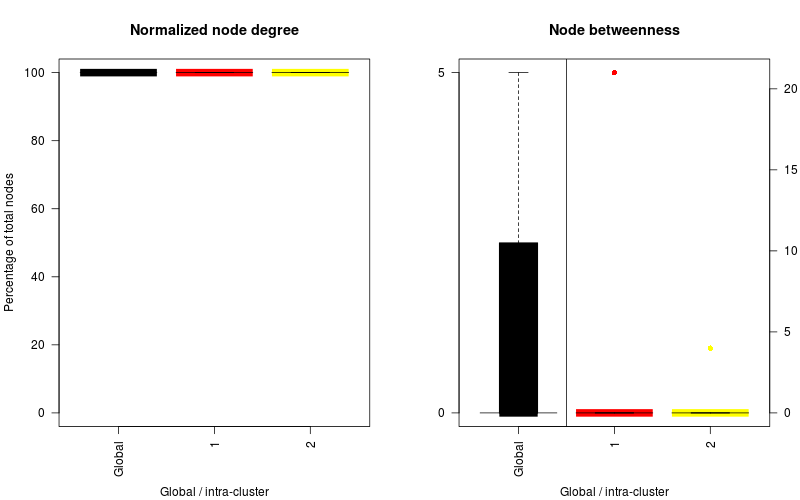
| Alcoholism | KEGG |
| Amine transport | GO |
| Amphetamine addiction | KEGG |
| Anatomical structure development * | GO |
| Anterograde trans-synaptic signaling | GO |
| Axon | GO |
| Axon part | GO |
| Axon terminus | GO |
| Binding | GO |
| Biological regulation | GO |
| Biosynthetic process | GO |
| Cell | GO |
| Cell body | GO |
| Cell-cell signaling | GO |
| Cell communication | GO |
| Cell part | GO |
| Cell periphery | GO |
| Cell projection | GO |
| Cell projection part | GO |
| Cellular localization | GO |
| Cellular metabolic process | GO |
| Cellular process | GO |
| Cellular response to stimulus * | GO |
| Chemical synaptic transmission | GO |
| Cocaine addiction | KEGG |
| Cytoplasm | GO |
| Cytoplasmic, membrane-bounded vesicle | GO |
| Cytoplasmic part | GO |
| Cytoplasmic vesicle | GO |
| Developmental process * | GO |
| Dopaminergic synapse | KEGG |
| Endomembrane system | GO |
| Establishment of localization | GO |
| Establishment of localization in cell | GO |
| Exocytic vesicle | GO |
| Extracellular region | GO |
| Integral component of membrane | GO |
| Integral component of plasma membrane | GO |
| Intracellular * | GO |
| Intracellular membrane-bounded organelle | GO |
| Intracellular organelle | GO |
| Intracellular part | GO |
| Intracellular vesicle | GO |
| Intrinsic component of membrane | GO |
| Intrinsic component of plasma membrane | GO |
| Localization | GO |
| Membrane | GO |
| Membrane-bounded organelle | GO |
| Membrane-bounded vesicle | GO |
| Membrane part | GO |
| Metabolic process | GO |
| Movement of cell or subcellular component | GO |
| Multicellular organismal process | GO |
| Multicellular organism development | GO |
| Negative regulation of biological process | GO |
| Negative regulation of cellular process | GO |
| Neuronal cell body | GO |
| Neuron part | GO |
| Neuron projection | GO |
| Neuron projection terminus | GO |
| Neurotransmitter transport | GO |
| Nitrogen compound transport | GO |
| Organelle | GO |
| Organic substance metabolic process | GO |
| Organic substance transport | GO |
| Plasma membrane | GO |
| Plasma membrane part | GO |
| Presynapse | GO |
| Primary metabolic process | GO |
| Protein binding | GO |
| Receptor binding | GO |
| Regulation of biological process | GO |
| Regulation of biological quality | GO |
| Regulation of cellular process | GO |
| Regulation of localization | GO |
| Regulation of multicellular organismal process | GO |
| Response to chemical | GO |
| Response to endogenous stimulus | GO |
| Response to nitrogen compound | GO |
| Response to organic substance | GO |
| Response to organonitrogen compound | GO |
| Response to stimulus | GO |
| Response to toxic substance | GO |
| Secretory vesicle | GO |
| Serotonergic synapse | KEGG |
| Signaling | GO |
| Single-multicellular organism process | GO |
| Single-organism cellular process | GO |
| Single-organism developmental process | GO |
| Single-organism localization | GO |
| Single-organism process | GO |
| Single organism signaling | GO |
| Single-organism transport | GO |
| Somatodendritic compartment | GO |
| Synapse | GO |
| Synapse part | GO |
| Synaptic signaling | GO |
| Synaptic vesicle | GO |
| System process | GO |
| Transmembrane transport | GO |
| Transport | GO |
| Transport vesicle | GO |
| Trans-synaptic signaling | GO |
| Vesicle | GO |
| |